SUIT-039 O2 mt D092: Difference between revisions
(Created page with "{{MitoPedia |abbr= |description=thumb|1D;2M.1;3Pal;3c;4M2;5P;6G;7S;8U;9Rot;10Ama.png |info='''A:''' Doerrier 2018 Met...") |
No edit summary |
||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{MitoPedia | {{MitoPedia | ||
|abbr= | |abbr= FAO and NS-pathways | ||
|description=[[File:1D;2M.1;3Pal;3c;4M2;5P;6G;7S;8U;9Rot;10Ama.png| | |description=[[File:1D;2M.1;3Pal;3c;4M2;5P;6G;7S;8U;9Rot;10Ama.png|400px]] | ||
|info='''A''' - '''[[SUIT-039]]''' - '''[[Fatty_acid_oxidation_pathway_control_state|F-pathway]] and [[NS-pathway]]''' | |||
|application=O2 | |||
|SUIT number=D092_1D;2M.1;3Pal;3c;4M2;5P;6G;7S;8U;9Rot;10Ama | |||
}} | }} | ||
{{Template:SUIT text D092}} | |||
__TOC__ | __TOC__ | ||
Communicated by [[ | Communicated by [[Grings M]], [[Cecatto C]] and [[Cardoso LHD]] (last update 2023-04-13) | ||
== | == Representative traces == | ||
{{Template:SUIT-039}} | |||
{{Template:SUIT- | |||
== Strengths and limitations == | == Strengths and limitations == | ||
:::* SUIT- | :::* SUIT-039 allows assessing FAO-linked respiration avoiding overestimation by endogenous substrates or malate anaplerosis. Other electron transfer pathways are also analyzed in OXPHOS (FN, FNS) and ET-state (FNS, S). | ||
:::+ SUIT- | :::+ SUIT-039 allows the depletion of endogenous substrates with ADP (1D). | ||
:::+ The protocol provides information on F-pathway in OXPHOS state. The low concentration of malate used in this protocol, typically 0.1 mM, does not saturate the N-pathway; but saturates the F-pathway. | :::+ The protocol provides information on F-pathway in OXPHOS state with palmitoylcarnitine, a long-chain acylcarnitine commonly found ''in vivo''. | ||
:::+ F-pathway ( | :::+ The low concentration of malate used in this protocol to assess F-pathway, typically 0.1 mM, does not saturate the N-pathway; but saturates the F-pathway. | ||
:::+ Pathway control in OXPHOS (F, F(N), FN, FNS | :::+ F-pathway (3Pal-2M.1) can be compared to FN-pathway (5P) in OXPHOS state. | ||
:::+ | :::+ Pathway control in OXPHOS (F, F(N), FN, FNS pathways) and in ET state (FNS and S) can be observed. | ||
:::+ Multiple pathways converging into Q (FNS) are assessed in OXPHOS and ET states. Therefore, ''P''/''E'' (7S/8U) at high ET capacity can be calculated. | |||
:::- | :::- LEAK state is not investigated. | ||
:::- | :::- Glycerophosphate (Gp) pathway is not investigated. | ||
== Compare SUIT protocols == | |||
::::* [[SUIT-036_O2_mt_D089]]: Similar protocol, including malate kinetics to assess mitochondrial malic enzyme and anaplerosis. | |||
::::* [[SUIT-002 O2 mt D005]]: Reference protocol SUIT protocol for isolated mitochondria, tissue homogenate and permeabilized cells (already permeabilized). Allows evaluation of FAO with octanoylcarnitine, also without overestimation by using low malate concentration. Besides this, provides information on pathway control in OXPHOS state (F, F(N), FN, FNS, FNSGp pathways) and ET-state (FNSGp, SGp pathways). | |||
::::* [[SUIT-025_O2_mt_D057]]: Allows evaluation of FAO with octanoylcarnitine, also without overestimation by using low malate concentration. Besides this, provides information on pathway control in OXPHOS state (F, F(N), FN, FNS pathways). | |||
::::* [[SUIT- | ::::* [[SUIT-041 O2 mt D096]]: A protocol to determine the optimum concentration of acylcarnitine. | ||
: | == Chemicals and syringes == | ||
::: | {{Template:Chemicals SUIT-039}} | ||
::: Suggested stock concentrations are shown in the specific DL-Protocol. | |||
== References == | |||
::: | <!-- | ||
: | == Download DatLab Protocol == | ||
::: | ::: DatLab Protocol (DLP) for DatLab 7.4: | ||
::: Excel analysis template (DatLab 7.4): | |||
--> | |||
<br /><br /> | |||
{{ | {{Support_FAT4BRAIN}} | ||
{{Labeling | |||
|additional=FAT4BRAINSUIT | |||
| | |||
}} | }} | ||
Latest revision as of 11:21, 14 April 2023
Description
Abbreviation: FAO and NS-pathways
Reference: A - SUIT-039 - F-pathway and NS-pathway
SUIT number: D092_1D;2M.1;3Pal;3c;4M2;5P;6G;7S;8U;9Rot;10Ama
O2k-Application: O2
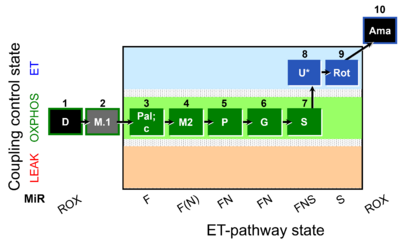
The SUIT-039 O2 mt D092 protocol provides a common reference for comparison of respiratory control of mitochondrial preparations such as isolated mitochondria, tissue homogenates and permeabilized cells (already permeabilized when they are added to the chamber) in a wide variety of species, tissues and cell types. SUIT-039 O2 mt D092 is specially designed to give information on F-pathway with palmitoylcarnitine as a FAO substrate in OXPHOS state avoiding FAO overestimation in the presence of anaplerotic pathways. Moreover, the pathway control in OXPHOS state (F, F(N), FN, FNS pathways) and in ET state (FNS and S) can be evaluated by using this SUIT protocol.
Communicated by Grings M, Cecatto C and Cardoso LHD (last update 2023-04-13)
Representative traces
Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 1D | ROX | 1D
| ||
| 2M.1 | 1D;2M.1 | |||
| 3Pal | PalMP | F | FAO | 1D;2M.1;3Pal
|
| 3c | PalMcP | F | FAO | 1D;2M.1;3Pal;3c
|
| 4M2 | PalMP | F(N) | FAO | 1D;2M.1;3Pal;3c;4M2
|
| 5P | PalPMP | FN | FAO&CI | 1D;2M.1;3Pal;4M2;5P
|
| 6G | PalPGMP | FN | FAO&CI | 1D;2M.1;3Pal;4M2;5P;6G
|
| 7S | PalPGMSP | FNS | FAO&CI&II | 1D;2M.1;3Pal;4M2;5P;6G;7S
|
| 8U | PalPGMSE | FNS | FAO&CI&II | 1D;2M.1;3Pal;4M2;5P;6G;7S;8U
|
| 10Rot | SE | S | CII | 1D;2M.1;3Pal;4M2;5P;6G;7S;8U;9Rot
|
| 11Ama | ROX | 1D;2M.1;3Pal;4M2;5P;6G;7S;8U;9Rot;10Ama
|
Strengths and limitations
- SUIT-039 allows assessing FAO-linked respiration avoiding overestimation by endogenous substrates or malate anaplerosis. Other electron transfer pathways are also analyzed in OXPHOS (FN, FNS) and ET-state (FNS, S).
- + SUIT-039 allows the depletion of endogenous substrates with ADP (1D).
- + The protocol provides information on F-pathway in OXPHOS state with palmitoylcarnitine, a long-chain acylcarnitine commonly found in vivo.
- + The low concentration of malate used in this protocol to assess F-pathway, typically 0.1 mM, does not saturate the N-pathway; but saturates the F-pathway.
- + F-pathway (3Pal-2M.1) can be compared to FN-pathway (5P) in OXPHOS state.
- + Pathway control in OXPHOS (F, F(N), FN, FNS pathways) and in ET state (FNS and S) can be observed.
- + Multiple pathways converging into Q (FNS) are assessed in OXPHOS and ET states. Therefore, P/E (7S/8U) at high ET capacity can be calculated.
- - LEAK state is not investigated.
- - Glycerophosphate (Gp) pathway is not investigated.
Compare SUIT protocols
- SUIT-036_O2_mt_D089: Similar protocol, including malate kinetics to assess mitochondrial malic enzyme and anaplerosis.
- SUIT-002 O2 mt D005: Reference protocol SUIT protocol for isolated mitochondria, tissue homogenate and permeabilized cells (already permeabilized). Allows evaluation of FAO with octanoylcarnitine, also without overestimation by using low malate concentration. Besides this, provides information on pathway control in OXPHOS state (F, F(N), FN, FNS, FNSGp pathways) and ET-state (FNSGp, SGp pathways).
- SUIT-025_O2_mt_D057: Allows evaluation of FAO with octanoylcarnitine, also without overestimation by using low malate concentration. Besides this, provides information on pathway control in OXPHOS state (F, F(N), FN, FNS pathways).
- SUIT-041 O2 mt D096: A protocol to determine the optimum concentration of acylcarnitine.
Chemicals and syringes
| Step | Chemical(s) and link(s) | Comments |
|---|---|---|
| 1D | ADP (D) | |
| 2M.1 | Malate (M) | |
| 3Pal | Palmitoylcarnitine (Pal) | |
| 3c | Cytochrome c (c) | |
| 4M2 | Malate (M) | |
| 5P | Pyruvate (P) | |
| 6G | Glutamate (G) | |
| 7S | Succinate (S) | |
| 8U | Carbonyl cyanide m-chlorophenyl hydrazone, CCCP (U) | Can be substituted for other uncoupler |
| 9Rot | Rotenone (Rot) | |
| 10Ama | Antimycin A (Ama) |
- Suggested stock concentrations are shown in the specific DL-Protocol.
References
- The project FAT4BRAIN has received funding from the European Union's Horizon 2020 research and innovation programme under grant agreement No 857394
Labels:
FAT4BRAINSUIT