|
|
| Line 2: |
Line 2: |
| |abbr=FNSGp(Oct,PGM) | | |abbr=FNSGp(Oct,PGM) |
| |description=[[File:1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot-.png|400px]] | | |description=[[File:1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot-.png|400px]] |
| | |info='''A''' Reference protocol RP2; pfi: permeabilized fibers - '''[[SUIT-002]]''' |
| | |application=O2 |
| |SUIT number=D006_1D;2M;3Oct;3c;4M;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd | | |SUIT number=D006_1D;2M;3Oct;3c;4M;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd |
| |info='''A: SUIT 2 is the RP2''' - [[SUIT-002]]
| |
| |application=O2
| |
| }} | | }} |
| ::: [[MitoPedia: SUIT]] - '''[[SUIT reference protocol]] RP2''' | | ::: [[MitoPedia: SUIT]] - '''[[SUIT reference protocol]] RP2 for permeabilized fibers''' |
| ::: '''[[Categories of SUIT protocols|SUIT-category]]:''' FNSGp(Oct,PGM) | | ::: '''[[Categories of SUIT protocols|SUIT-category]]:''' FNSGp(Oct,PGM) |
| ::: '''[[SUIT protocol pattern]]:''' diametral 1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot | | ::: '''[[SUIT protocol pattern]]:''' diametral 1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot |
|
| |
| == Harmonized SUIT protocols ==
| |
|
| |
|
| |
| === RP1 and RP2 ===
| |
| '''SUIT RP1''' [[File:1PM;2D;2c;3U;4G;5S;6Oct;7Rot;8Gp;9Ama;10AsTm;11Azd.png|400px|SUIT-RP1]]
| |
| '''SUIT RP2''' [[File:1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd.png|500px|SUIT-RP2]]
| |
|
| |
| :::: [[1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp-|SUIT RP1]]: 1PM;2D;2c;3U;4G;5S;6Oct;7Rot;'''8Gp''';9Ama;'''10AsTm''';11Azd
| |
| :::: '''SUIT RP2''': 1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;'''10Rot''';11Ama;'''12AsTm''';13Azd
| |
|
| |
| [[File:RP1&RP2.png|400px]] '''Harmonization between RP1 and RP2'''
| |
|
| |
| === RP1 and RP2 in cells ===
| |
| RP2 can be assessed in cells after the selective plasma membrane permeabilization. Digitonin is a mild detergent that permeabilizes plasma membranes. The optimum effective digitonin concentration for complete plasma membrane permeabilization of cultured cells can be determined directly in a respirometric protocol (see [[Digitonin]]). The use of intact cells allows the possibility to evaluate respiration in two different modules: intact cells and permeabilized cells. ROUTINE respiration (ce1) is obtained before digitonin is employed, whereas after the use of digitonin we can address differentet [[Electron transfer-pathway state|Electron transfer-pathway statea]].
| |
|
| |
| :::: [[1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp-#Harmonized_SUIT_protocols|SUIT RP1 in cells]]: '''ce1''';1Dig;1PM;2D;2c;3U;4G;5S;6Oct;7Rot;'''8Gp''';9Ama;'''10Tm''';11Azd
| |
| :::: '''SUIT RP2 in cells''': '''ce1''';1Dig;1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;'''10Rot''';11Ama;'''12AsTm''';13Azdd
| |
|
| |
| [[File:RP1&RP2_pce(Dig).png|400px]] '''Harmonization between RP1 and RP2 in cells'''
| |
|
| |
|
| |
| === RP1 and RP2 in PBMCs and PTLs ===
| |
| In PBMCs and PLTs the reference protocol RP1 is performed in the absense of Oct: [[1PM;2D;3U;4G;5S;6Rot;7Gp-|SUIT 1 for PBMCs and PTLs]]
| |
|
| |
| :::: [[1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp-#Harmonized_SUIT_protocols|SUIT RP1 in PBMCs and PTLs]]'''SUIT RP1 for PBMCs and PLTs''':1PM;2D;2c;3U;4G;5S;6Rot;'''7Gp''';8Ama;'''9AsTm''';10Azd
| |
| :::: '''SUIT RP2 PBMCs and PLTs''': 1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;'''10Rot''';11Ama;'''12AsTm''';13Azd
| |
|
| |
| [[File:Reference protocol RP1&RP2_pce_PBMCs_PTLs.png|400px]] '''Harmonization between RP1 and RP2 in PBMC and PTL cells'''
| |
|
| |
|
| |
|
| == References == | | == References == |
| Line 54: |
Line 24: |
|
| |
|
| {{Template:SUIT-002}} | | {{Template:SUIT-002}} |
| | |
| | == Strengths and limitations == |
| | |
| | == Compare SUIT protocols == |
| | |
| | ::::* [[SUIT-001 O2 pfi D002]] (RP1): Harmonized SUIT protocol for permeabilized fibers. |
|
| |
|
| {{MitoPedia concepts | | {{MitoPedia concepts |
| Line 61: |
Line 37: |
| |mitopedia method=Respirometry | | |mitopedia method=Respirometry |
| }} | | }} |
| | {{Labeling |
| | |additional=SUIT-001}} |
| | [[Category:Publications]] |

- high-resolution terminology - matching measurements at high-resolution
SUIT-002 O2 pfi D006
Description
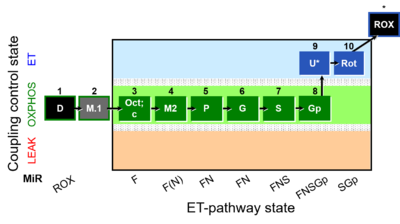
Abbreviation: FNSGp(Oct,PGM)
Reference: A Reference protocol RP2; pfi: permeabilized fibers - SUIT-002
SUIT number: D006_1D;2M;3Oct;3c;4M;5P;6G;7S;8Gp;9U;10Rot;11Ama;12AsTm;13Azd
O2k-Application: O2
- MitoPedia: SUIT - SUIT reference protocol RP2 for permeabilized fibers
- SUIT-category: FNSGp(Oct,PGM)
- SUIT protocol pattern: diametral 1D;2M.1;3Oct;3c;4M2;5P;6G;7S;8Gp;9U;10Rot
References
Steps and respiratory states
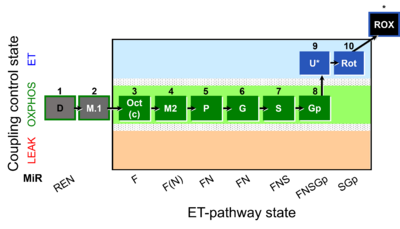
| Step
|
State
|
Pathway
|
Q-junction
|
Comment - Events (E) and Marks (M)
|
| 1D
|
REN
|
|
|
1D
- ADP is added to stimulate the consumption of endogenous fuel-substrates.
|
| 2M.1
|
|
|
|
1D;2M.1
|
| 3Oct
|
OctMP
|
F
|
FAO
|
1D;2M.1;3Oct
- Respiratory stimulation of the FAO-pathway, F, by fatty acid, FA, in the presence of malate, M. Malate is a type N substrate (N), required for the F-pathway. The FA concentration has to be optimized to saturate the F-pathway, without inhibiting or uncoupling respiration.
|
| 3c
|
OctMcP
|
F
|
FAO
|
1D;2M.1;3Oct;3c
- Respiratory stimulation of the FAO-pathway, F, by fatty acid, FA, in the presence of malate, M. Malate is a type N substrate (N), required for the F-pathway. The FA concentration has to be optimized to saturate the F-pathway, without inhibiting or uncoupling respiration.
- OXPHOS capacity P (with saturating [ADP]), active OXPHOS state.
- Addition of cytochrome c yields a test for integrity of the mtOM (cytochrome c control efficiency). Stimulation by added cytochrome c would indicate an injury of the mtOM and limitation of respiration in the preceding state without added c due to loss of cytochrome c. Typically, cytochrome c is added immediately after the earliest ADP-activation step (OXPHOS capacity P with saturating [ADP]).
|
| 4M2
|
OctMP
|
F(N)
|
FAO
|
1D;2M.1;3Oct;3c;4M2
- Respiratory stimulation of the FAO-pathway, F, by fatty acid FA in the presence of malate M. Malate is a type N substrate (N), required for the F-pathway. In the presence of anaplerotic pathways (e.g., mitochondrial malic enzyme, mtME) the F-pathway capacity is overestimated, if there is an added contribution of NADH-linked respiration, F(N) (see SUIT-002). The FA concentration has to be optimized to saturate the FAO-pathway, without inhibiting or uncoupling respiration.
- High concentration of malate, typically 2 mM, saturates the N-pathway.
- OXPHOS capacity P (with saturating [ADP]), active OXPHOS state.
|
| 5P
|
OctPMP
|
FN
|
FAO&CI
|
1D;2M.1;3Oct;4M2;5P
- Respiratory stimulation by simultaneous action of the F-pathway and N-pathway with convergent electron flow in the FN-pathway for evaluation of an additive or inhibitory effect of F.
- OXPHOS capacity P (with saturating [ADP]), active OXPHOS state.
|
| 6G
|
OctPGMP
|
FN
|
FAO&CI
|
1D;2M.1;3Oct;4M2;5P;6G
- Respiratory stimulation by simultaneous action of the F-pathway and N-pathway with convergent electron flow in the FN-pathway for evaluation of an additive or inhibitory effect of F.
- OXPHOS capacity P (with saturating [ADP]), active OXPHOS state.
|
| 7S
|
OctPGMSP
|
FNS
|
FAO&CI&II
|
1D;2M.1;3Oct;4M2;5P;6G;7S
- Respiratory stimulation by simultaneous action of the F-pathway, N-pathway, and S-pathway, with convergent electron flow in the FNS-pathway for reconstitution of TCA cycle function and additive or inhibitory effect of F.
- OXPHOS capacity P (with saturating [ADP]), active OXPHOS state.
|
| 8Gp
|
OctPGMSGpP
|
FNSGp
|
FAO&CI&II&GpDH
|
1D;2M.1;3Oct;4M2;5P;6G;7S;8Gp
|
| 9U
|
OctPGMSGpE
|
FNSGp
|
FAO&CI&II&GpDH
|
1D;2M.1;3Oct;4M2;5P;6G;7S;8Gp;9U
|
| 10Rot
|
SGpE
|
SGp
|
CII&GpDH
|
1D;2M.1;3Oct;4M2;5P;6G;7S;8Gp;9U;10Rot
- Respiratory stimulation by action of succinate and glycerophosphate, Gp, with convergent electron flow in the SGp-pathway (CII&GpDH-linked pathway to the Q-junction).
- Noncoupled electron transfer state, ET state, with ET capacity E.
|
| 11Ama
|
ROX
|
|
|
1D;2M.1;3Oct;4M2;5P;6G;7S;8Gp;9U;10Rot;11Ama
- Rox is the residual oxygen consumption in the ROX state, due to oxidative side reactions, estimated after addition of antimycin A (inhibitor of CIII). Rox is subtracted from oxygen flux as a baseline for all respiratory states, to obtain mitochondrial respiration (mt).
|

- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- » Coupling control state
- » ET capacity
- » OXPHOS capacity
- » LEAK respiration
- Pathway control
- » Electron transfer pathway
- » Fatty acid oxidation pathway control state, F
- » NADH electron transfer-pathway state, N
- » Succinate pathway control state, S
- » NS-pathway control state, NS
- » Glycerophosphate pathway control state, Gp
- » Complex IV single step, CIV
- » Anaplerotic pathway control state
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Glossary
- » List of SUIT states
- » SUIT concept
Strengths and limitations
Compare SUIT protocols
MitoPedia concepts:
SUIT protocol,
SUIT A,
Find
MitoPedia methods:
Respirometry
Labels:
SUIT-001




