Difference between revisions of "PM-pathway control state"
| Line 7: | Line 7: | ||
'''SUIT protocol:''' [[SUIT-001|1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp-]] - SUIT_RP1 | '''SUIT protocol:''' [[SUIT-001|1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp-]] - SUIT_RP1 | ||
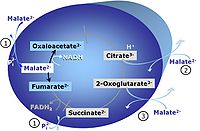
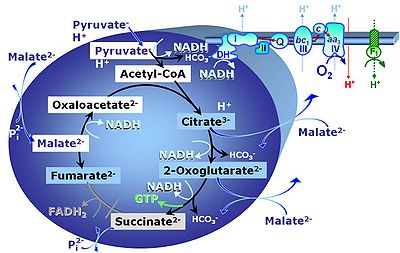
[[Pyruvate]] (P) is oxidatively decarboxylated to acetyl-CoA and CO<sub>2</sub>, yielding [[NADH]] catalyzed by pyruvate dehydrogenase. [[Malate]] (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). 2-oxoglutarate (α-ketoglutarate) is formed from isocitrate (isocitrate dehydrogenase). | Upstream of the NAD-junction, [[Pyruvate]] (P) is oxidatively decarboxylated to acetyl-CoA and CO<sub>2</sub>, yielding [[NADH]] catalyzed by pyruvate dehydrogenase. [[Malate]] (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). 2-oxoglutarate (α-ketoglutarate) is formed from isocitrate (isocitrate dehydrogenase). | ||
|info=[[Gnaiger 2020 BEC MitoPathways]] | |info=[[Gnaiger 2020 BEC MitoPathways]] | ||
}} | }} | ||
__TOC__ | |||
[[File:PM.jpg|right|400px|link=Gnaiger 2020 BEC MitoPathways |Gnaiger 2020 BEC MitoPathways]] | [[File:PM.jpg|right|400px|link=Gnaiger 2020 BEC MitoPathways |Gnaiger 2020 BEC MitoPathways]] | ||
== PM<sub>''L''</sub> == | == PM<sub>''L''</sub> == | ||
LEAK | With PM as N-substrates, LEAK respiration ''L'' can be evaluated in the following SUIT protocols: | ||
:::*[[SUIT-001]] | :::*[[SUIT-001]] | ||
::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-001 O2 mt D001]] | ::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-001 O2 mt D001]] | ||
| Line 32: | Line 30: | ||
== PM<sub>''P''</sub> == | == PM<sub>''P''</sub> == | ||
OXPHOS | With PM as N-substrates, OXPHOS capacity ''P'' can be evaluated in the following SUIT protocols: | ||
:::*[[SUIT-001]] | :::*[[SUIT-001]] | ||
::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-001 O2 mt D001]] | ::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-001 O2 mt D001]] | ||
| Line 47: | Line 45: | ||
== PM<sub>''E''</sub> == | == PM<sub>''E''</sub> == | ||
ET | With PM as N-substrates, ET capacity ''E'' can be evaluated in the following SUIT protocols: | ||
:::*[[SUIT-001]] | :::*[[SUIT-001]] | ||
::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-001 O2 mt D001]] | ::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-001 O2 mt D001]] | ||
| Line 62: | Line 60: | ||
== Linear coupling control in the N-pathway control state: ''L | == Linear coupling control in the N-pathway control state: ''L → P → E'' == | ||
:::* '''''P-L''''' | |||
:::: [[ | :::: [[P-L control efficiency |''P-L'' control efficiency]], ''j<sub>P-L</sub>'' = (''P-L'')/''P'' = 1-''L/P'', is measured in the N-pathway state, with defined coupling sites (CI, CIII, CIV). | ||
:::* '''''P-E''''' | |||
:::: [[CCCP]] is titrated stepwise to maximum flux, to evaluate limitation of OXPHOS by the phosphorylation system, expressed as the | :::: [[CCCP]] is titrated stepwise to maximum flux, to evaluate limitation of OXPHOS by the phosphorylation system, expressed as the [[E-P control efficiency |''E-P'' control efficiency]] ''j<sub>E-P</sub>'' = (''E-P'')/''E'' = 1-''P/E''. | ||
:::: If ''j<sub>E-P</sub>''>0, then the [[E-L coupling efficiency]] rather than the [[P-L control efficiency |''P-L'' control efficiency]] is the proper expression of coupling, ''j<sub>E-L</sub>'' = (''E-L'')/''E'' = 1-''L/E''. | |||
| Line 74: | Line 73: | ||
:::: The [[Pyruvate anaplerotic pathway control state]] (pyruvate alone) is not an ET-pathway competent substrate state in most mt-preparations, since acetyl-CoA accumulates without the co-substrate (oxaloacetate) of citrate synthase. | :::: The [[Pyruvate anaplerotic pathway control state]] (pyruvate alone) is not an ET-pathway competent substrate state in most mt-preparations, since acetyl-CoA accumulates without the co-substrate (oxaloacetate) of citrate synthase. | ||
:::: The [[Malate-anaplerotic pathway control state]] (M alone) is not an ET-pathway competent substrate state in many mt-preparations, since oxaloacetate accumulates without the co-substrate (acetyl-CoA) of citrate synthase. | :::: The [[Malate-anaplerotic pathway control state]] (M alone) is not an ET-pathway competent substrate state in many mt-preparations, since oxaloacetate accumulates without the co-substrate (acetyl-CoA) of citrate synthase. | ||
{{MitoPedia concepts | |||
|mitopedia concept=SUIT state | |||
}} | |||
Revision as of 23:22, 23 May 2022
Description
MitoPathway control state: NADH Electron transfer-pathway state
SUIT protocol: 1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp- - SUIT_RP1
Upstream of the NAD-junction, Pyruvate (P) is oxidatively decarboxylated to acetyl-CoA and CO2, yielding NADH catalyzed by pyruvate dehydrogenase. Malate (M) is oxidized to oxaloacetate by mt-malate dehydrogenase located in the mitochondrial matrix. Condensation of oxaloacate with acetyl-CoA yields citrate (citrate synthase). 2-oxoglutarate (α-ketoglutarate) is formed from isocitrate (isocitrate dehydrogenase).
Abbreviation: PM
Reference: Gnaiger 2020 BEC MitoPathways
PML
With PM as N-substrates, LEAK respiration L can be evaluated in the following SUIT protocols:
-
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-001 O2 mt D001
- DL-Protocol for permeabilized fibers (pfi): SUIT-001 O2 pfi D002
- DL-Protocol for permeabilized cells (pce): SUIT-001 O2 ce-pce D003
- DL-Protocol for permeabilized blood cells (PBMC and PLT): SUIT-001 O2 ce-pce D004
- DL-Protocol for permeabilized fibers (pfi): SUIT-004 O2 pfi D010
- DL-Protocol for permeabilized cells (pce): SUIT-006 O2 ce-pce D029
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-006 O2 mt D047
- DL-Protocol for permeabilized fibers (pfi): SUIT-008 O2 pfi D014
-
PMP
With PM as N-substrates, OXPHOS capacity P can be evaluated in the following SUIT protocols:
-
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-001 O2 mt D001
- DL-Protocol for permeabilized fibers (pfi): SUIT-001 O2 pfi D002
- DL-Protocol for permeabilized cells (pce): SUIT-001 O2 ce-pce D003
- DL-Protocol for permeabilized blood cells (PBMC and PLT): SUIT-001 O2 ce-pce D004
- DL-Protocol for permeabilized fibers (pfi): SUIT-004 O2 pfi D010
- DL-Protocol for permeabilized cells (pce): SUIT-006 O2 ce-pce D029
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-006 O2 mt D047
- DL-Protocol for permeabilized fibers (pfi): SUIT-008 O2 pfi D014
-
PME
With PM as N-substrates, ET capacity E can be evaluated in the following SUIT protocols:
-
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-001 O2 mt D001
- DL-Protocol for permeabilized fibers (pfi): SUIT-001 O2 pfi D002
- DL-Protocol for permeabilized cells (pce): SUIT-001 O2 ce-pce D003
- DL-Protocol for permeabilized blood cells (PBMC and PLT): SUIT-001 O2 ce-pce D004
- DL-Protocol for permeabilized fibers (pfi): SUIT-004 O2 pfi D010
- DL-Protocol for permeabilized cells (pce): SUIT-006 O2 ce-pce D029
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-006 O2 mt D047
- DL-Protocol for permeabilized fibers (pfi): SUIT-008 O2 pfi D014
-
Linear coupling control in the N-pathway control state: L → P → E
- P-L
- P-L control efficiency, jP-L = (P-L)/P = 1-L/P, is measured in the N-pathway state, with defined coupling sites (CI, CIII, CIV).
- P-E
- CCCP is titrated stepwise to maximum flux, to evaluate limitation of OXPHOS by the phosphorylation system, expressed as the E-P control efficiency jE-P = (E-P)/E = 1-P/E.
- If jE-P>0, then the E-L coupling efficiency rather than the P-L control efficiency is the proper expression of coupling, jE-L = (E-L)/E = 1-L/E.
Discussion
- The Pyruvate anaplerotic pathway control state (pyruvate alone) is not an ET-pathway competent substrate state in most mt-preparations, since acetyl-CoA accumulates without the co-substrate (oxaloacetate) of citrate synthase.
- The Malate-anaplerotic pathway control state (M alone) is not an ET-pathway competent substrate state in many mt-preparations, since oxaloacetate accumulates without the co-substrate (acetyl-CoA) of citrate synthase.
MitoPedia concepts:
SUIT state