Difference between revisions of "MitoEAGLE protocols, terminology, documentation"
From Bioblast
Nirschl Lisa (talk | contribs) |
|||
| (30 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
__TOC__ | __TOC__ | ||
<br /> | <br /> | ||
<big><big><big>'''WG1''' | <big><big><big>'''WG1 Project application''' | ||
:::: '''Standard operating procedures and user requirement document: Protocols, terminology, documentation''' | :::: '''Standard operating procedures and user requirement document: Protocols, terminology, documentation''' | ||
</big></big></big> | </big></big></big> | ||
::::» [[WG1 MitoEAGLE protocols, terminology, documentation |WG1 Action]] | |||
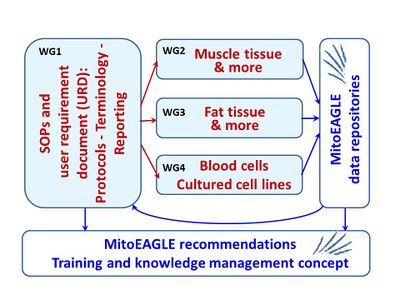
[[File:MITOEAGLE Working groups.jpg|right|400px|MitoEAGLE Working Groups]] | |||
[[File:MITOEAGLE Working groups.jpg|right|400px| | |||
:::: The quality management system, scope of SOPs to be elaborated and the outline of the data repositories will be defined in a user requirement document. | :::: The quality management system, scope of SOPs to be elaborated and the outline of the data repositories will be defined in a user requirement document. | ||
| Line 14: | Line 13: | ||
=== Respirometric reference protocols === | === Respirometric reference protocols === | ||
:::: A ‘library of protocols’ applied in mitochondrial respiratory physiology will be collected in a standard format, delineating the diversity of experimental approaches to assess bioenergetic function. A set of reference protocols will rigorously document (Maelstrom Research program) the state-of-the-art standards in designing, conducting, reporting, interpreting, and validating such bioenergetic tests (see also | :::: A ‘library of protocols’ applied in mitochondrial respiratory physiology will be collected in a standard format, delineating the diversity of experimental approaches to assess bioenergetic function. A set of reference protocols will rigorously document (Maelstrom Research program) the state-of-the-art standards in designing, conducting, reporting, interpreting, and validating such bioenergetic tests (see also figure on the right). | ||
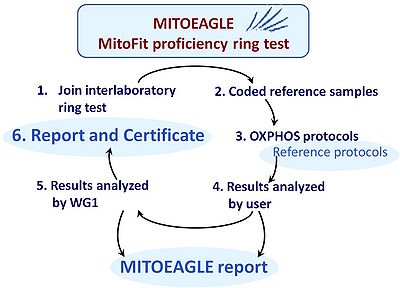
[[File:MITOEAGLE Proficiency ring test.jpg|right|400px| | [[File:MITOEAGLE Proficiency ring test.jpg|right|400px|MitoEAGLE Proficiency ring test]] | ||
=== Interlaboratory proficiency test === | === Interlaboratory proficiency test === | ||
:::: Inter-laboratory ring tests are a basic requirement for quality management. Such proficiency tests require a reference sample which must be homogenous, stable, representative of the diagnostic target, amenable for distribution, and economical for large-scale production. For respiratory OXPHOS analysis a reference sample of structurally and functionally intact mitochondria is not yet available to perform corresponding ring tests (in contrast to enzymatic OXPHOS ring tests). Recently, the widely applied human cell line HEK 293T was found to be potentially suitable for cryopreservation. Upon thawing the cells can be used immediately for respirometry of both intact and permeabilized cells. Thus interlaboratory proficiency testing may now be feasible as a world-wide innovation in the field of mitochondrial respiratory physiology. Participating labs may implement the test for intra- and interlaboratory validation and longitudinal performance monitoring, using a reference OXPHOS protocol (Fig. 3). The organization of the proficiency test should follow the requirements of the [[ISO/IEC 17043:2010 General requirements for proficiency testing |ISO 17043 and 13528]] standards. A corresponding | :::: Inter-laboratory ring tests are a basic requirement for quality management. Such proficiency tests require a reference sample which must be homogenous, stable, representative of the diagnostic target, amenable for distribution, and economical for large-scale production. For respiratory OXPHOS analysis a reference sample of structurally and functionally intact mitochondria is not yet available to perform corresponding ring tests (in contrast to enzymatic OXPHOS ring tests). Recently, the widely applied human cell line HEK 293T was found to be potentially suitable for cryopreservation. Upon thawing the cells can be used immediately for respirometry of both intact and permeabilized cells. Thus interlaboratory proficiency testing may now be feasible as a world-wide innovation in the field of mitochondrial respiratory physiology. Participating labs may implement the test for intra- and interlaboratory validation and longitudinal performance monitoring, using a reference OXPHOS protocol (Fig. 3). The organization of the proficiency test should follow the requirements of the [[ISO/IEC 17043:2010 General requirements for proficiency testing |ISO 17043 and 13528]] standards. A corresponding MitoEAGLE proficiency training shall be developed and implemented. Feedback from the participants will lead to a final adjustment of the SOPs in the proficiency test, which will be made generally available. | ||
=== Instrumental platforms === | === Instrumental platforms === | ||
| Line 24: | Line 23: | ||
=== Nomenclature === | === Nomenclature === | ||
:::: Harmonization of nomenclature on mitochondrial respiratory states and control parameters: The logistics of development of a database requires application of strictly defined terms for all included variables. There is no general reference available upon which a consistent terminology on mitochondrial physiology and bioenergetics can be based. The | :::: Harmonization of nomenclature on mitochondrial respiratory states and control parameters: The logistics of development of a database requires application of strictly defined terms for all included variables. There is no general reference available upon which a consistent terminology on mitochondrial physiology and bioenergetics can be based. The MitoEAGLE consortium, therefore, has to accomplish the ambitious goal to unify and simplify the terminology in the field for the purpose of the QMS, which will lead to the development of recommendations for the use of a common terminology in mitochondrial physiological research. A publication will be prepared as an Open Access article which will be a milestone towards a unification of concepts and nomenclature. | ||
=== Knowledge management system === | === Knowledge management system === | ||
| Line 30: | Line 29: | ||
:::: With an increase in data volume and complexity comes along the requirement for standardized report formats and digital accessibility of the data. Since such a database is still missing for mitochondrial physiology, the goal of WG1 is to establish an integrated and analytic environment linking mitochondrial respiration with external data (cell physiology, anthropometry, spiroergometry, lifestyle, genomics) in the context of physical fitness evaluation and clinical diagnostics. Databases will be established combining biological and clinical data comprising proprietary and publicly available datasets into one integrated solution. Data import and export functionalities will allow data upload and mapping to corresponding objects in the database, in this way generating valuable data and allowing for project-specific as well as cross-project data mining. | :::: With an increase in data volume and complexity comes along the requirement for standardized report formats and digital accessibility of the data. Since such a database is still missing for mitochondrial physiology, the goal of WG1 is to establish an integrated and analytic environment linking mitochondrial respiration with external data (cell physiology, anthropometry, spiroergometry, lifestyle, genomics) in the context of physical fitness evaluation and clinical diagnostics. Databases will be established combining biological and clinical data comprising proprietary and publicly available datasets into one integrated solution. Data import and export functionalities will allow data upload and mapping to corresponding objects in the database, in this way generating valuable data and allowing for project-specific as well as cross-project data mining. | ||
:::: The ability to formulate meta-studies in our approach is an important success factor: The platform can aggregate the project’s complete knowledge and provides the ability to make diagnostic profiling associated with diseases. In addition to functional data, other data types such as nutritional or environmental factors associated with disease prevention can be identified and aggregated into the | :::: The ability to formulate meta-studies in our approach is an important success factor: The platform can aggregate the project’s complete knowledge and provides the ability to make diagnostic profiling associated with diseases. In addition to functional data, other data types such as nutritional or environmental factors associated with disease prevention can be identified and aggregated into the MitoEAGLE Knowledge Management System. | ||
:::: Querying the heterogeneous data types and result representation are the most important requirements of the knowledge platform besides to data integration and modelling. The application will feature different types of visualization techniques for all integrated data types. The database will be developed using the concept of multi-tier client-server architecture. Individual components are separated corresponding to their functionality. An upstream web server handles user requests, an application server performing business logic, and a back-end database system guaranties persistent data storage. All components are additionally separated by means of network security incorporating firewalls and are hosted in a secure environment. The platform administration ensures privacy regarding proprietary data by user rights policies. | :::: Querying the heterogeneous data types and result representation are the most important requirements of the knowledge platform besides to data integration and modelling. The application will feature different types of visualization techniques for all integrated data types. The database will be developed using the concept of multi-tier client-server architecture. Individual components are separated corresponding to their functionality. An upstream web server handles user requests, an application server performing business logic, and a back-end database system guaranties persistent data storage. All components are additionally separated by means of network security incorporating firewalls and are hosted in a secure environment. The platform administration ensures privacy regarding proprietary data by user rights policies. | ||
:::: It will be the task of WG1 to continuously collect the input from all other WGs, compare, discuss and align SOPs as far as possible, and to finally develop recommendations for quality control, data reporting and data sharing beyond the published record. In addition, WG1 will develop concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation. Finally, it will provide a summary on strategic dissemination and an education programme for | :::: It will be the task of WG1 to continuously collect the input from all other WGs, compare, discuss and align SOPs as far as possible, and to finally develop recommendations for quality control, data reporting and data sharing beyond the published record. In addition, WG1 will develop concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation. Finally, it will provide a summary on strategic dissemination and an education programme for MitoEAGLE. The education programme will ensure that not only presently participating groups will adhere to the set standards but that also groups joining in at later points may smoothly fit into the community and help to further broaden and deepen our understanding on mitochondrial physiology in health and disease. | ||
== | == Management == | ||
=== Tasks === | === Tasks === | ||
:::# Opening of the database to the research public for data search and moderated entry of new | :::# Opening of the database to the research public for data search and moderated entry of new datasets. | ||
:::# Development of interlaboratory proficiency ring test, of recommendation/certification reports and of a | :::# Development of interlaboratory proficiency ring test, of recommendation/certification reports and of a MitoEAGLE proficiency training module. | ||
:::# Sample distribution, ring test experiments ( | :::# Sample distribution, ring test experiments (MitoEAGLE-proficiency training on two different points of time, 6 months apart), data analysis by experimenters and MitoEAGLE-proficiency test manager. | ||
:::# Reports and publication: Reports with specific results in comparison with an anonymous data summary, and joint publication. Individual reports may be complemented by recommendations, and are summarized in an informal certification report | :::# Reports and publication: Reports with specific results in comparison with an anonymous data summary, and joint publication. Individual reports may be complemented by recommendations, and are summarized in an informal certification report. | ||
:::# Instrumental platform comparison with reference sample applying comparable measurements. | :::# Instrumental platform comparison with reference sample applying comparable measurements. | ||
:::# Draft, dissemination, collection and final formulation of a manuscript on concepts and terminology of mitochondrial physiology | :::# Draft, dissemination, collection and final formulation of a manuscript on concepts and terminology of mitochondrial physiology. | ||
:::# Collect and discuss procedures and experimental protocols for the evaluation of mitochondrial capacities and create a library of protocols: A database comprising reference SUIT protocols, standard experimental media, and detailed instructions for OXPHOS analysis is assembled. | :::# Collect and discuss procedures and experimental protocols for the evaluation of mitochondrial capacities and create a library of protocols: A database comprising reference SUIT protocols, standard experimental media, and detailed instructions for OXPHOS analysis is assembled. | ||
:::# | :::# MitoEAGLE data management system (DMS) development: The DMS for further data preprocessing. The DMS will enable management and analysis of instrument-specific and project-specific data. | ||
:::# | :::# MitoEAGLE database (DB) development: DB will facilitate high-level project specific analysis as well as meta-analyses across projects, visualization of project specific data as well as integrative analyses of datasets for various proprietary and/or publicly available datasets. | ||
:::# Testing of developed software components regarding functionality and usability, modification and implementation. Deployment for routine testing in the labs of our strategic partners. | :::# Testing of developed software components regarding functionality and usability, modification and implementation. Deployment for routine testing in the labs of our strategic partners. | ||
:::# Data input: Upload of datasets generated through WG 2-4 in internal lab developments, results of the proficiency ring test, and during application module developments. In addition, the database will be populated with available proprietary and public datasets. | :::# Data input: Upload of datasets generated through WG 2-4 in internal lab developments, results of the proficiency ring test, and during application module developments. In addition, the database will be populated with available proprietary and public datasets. | ||
:::# Develop recommendations for quality control, data reporting and data sharing beyond the published record. Develop concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation | :::# Develop recommendations for quality control, data reporting and data sharing beyond the published record. Develop concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation. | ||
:::# Provide a summary on strategic dissemination and an education programme for | :::# Provide a summary on strategic dissemination and an education programme for MitoEAGLE. | ||
| Line 69: | Line 55: | ||
:::# Data sets entered in standardized format and database opened for the public | :::# Data sets entered in standardized format and database opened for the public | ||
:::# Protocols for ring-tests and standard analysis and report scheme defined | :::# Protocols for ring-tests and standard analysis and report scheme defined | ||
:::# | :::# MitoEAGLE proficiency training workshop finished | ||
:::# Analysis is completed and joint publication submitted | :::# Analysis is completed and joint publication submitted | ||
:::# Publication of paper on concepts and terminology | :::# Publication of paper on concepts and terminology | ||
:::# Library of protocols online | :::# Library of protocols online | ||
:::# Publication of recommended protocols and procedures | :::# Publication of recommended protocols and procedures | ||
:::# Guidelines for acquisition, evaluation and data documentation available | :::# Guidelines for acquisition, evaluation and data documentation available | ||
:::# | :::# MitoEAGLE data management system finished | ||
:::# | :::# MitoEAGLE database ready for use | ||
:::# Software modifications incorporated following extensive tests | :::# Software modifications incorporated following extensive tests | ||
:::# Publication of | :::# Publication of MitoEAGLE-KMP | ||
:::# Recommendations on data acquisition, evaluation and management published | :::# Recommendations on data acquisition, evaluation and management published | ||
:::# Concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation published | :::# Concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation published | ||
=== Deliverables === | === Deliverables === | ||
:::# Guidelines for future research and recommendations for the evaluation of respiratory characteristics in human respirometric samples | :::# Guidelines for future research and recommendations for the evaluation of respiratory characteristics in human respirometric samples | ||
:::# A | :::# A database on mt fitness evaluated with human cells and tissues | ||
:::# Training of researchers towards improved reproducibility of sample preparation and respirometric evaluation | :::# Training of researchers towards improved reproducibility of sample preparation and respirometric evaluation | ||
:::# Qualitative and quantitative evaluation of results obtained in ongoing studies by comparison with a reference sample | :::# Qualitative and quantitative evaluation of results obtained in ongoing studies by comparison with a reference sample | ||
:::# An expanded | :::# An expanded MitoEAGLE database | ||
:::# A joint publication documenting the results of the | :::# A joint publication documenting the results of the MitoEAGLE-PT | ||
:::# A multi-authored publication towards a unification of concepts and nomenclature in mitochondrial physiology | :::# A multi-authored publication towards a unification of concepts and nomenclature in mitochondrial physiology | ||
:::# A library of protocols for sample preparation and examination by respirometry | :::# A library of protocols for sample preparation and examination by respirometry | ||
:::# A multi-authored publication with recommendations of comparable standard protocols and procedures | :::# A multi-authored publication with recommendations of comparable standard protocols and procedures. | ||
:::# Guidelines as to the optimum use of a reference sample for determination of respiration and related parameters of interest | :::# Guidelines as to the optimum use of a reference sample for determination of respiration and related parameters of interest. | ||
:::# The | :::# The MitoEAGLE data management system. | ||
:::# A continuously updated | :::# A continuously updated MitoEAGLE database for the use as reference data and guideline towards application of optimized protocols, ultimately leading towards standardization. | ||
:::# A joint publication on the use of the | :::# A joint publication on the use of the MitoEAGLE database and management system. | ||
:::# An education programme for the use of | :::# An education programme for the use of MitoEAGLE database and management system. | ||
{{MITOEAGLE banner}} | |||
Latest revision as of 12:05, 19 January 2018
| News and Events | Working Groups | Short-Term Scientific Missions | Management Committee | Members |
COST Action CA15203 (2016-2021): MitoEAGLE
Evolution-Age-Gender-Lifestyle-Environment: mitochondrial fitness mapping
MitoEAGLE protocols, terminology, documentation
WG1 Project application
- Standard operating procedures and user requirement document: Protocols, terminology, documentation
- The quality management system, scope of SOPs to be elaborated and the outline of the data repositories will be defined in a user requirement document.
Respirometric reference protocols
- A ‘library of protocols’ applied in mitochondrial respiratory physiology will be collected in a standard format, delineating the diversity of experimental approaches to assess bioenergetic function. A set of reference protocols will rigorously document (Maelstrom Research program) the state-of-the-art standards in designing, conducting, reporting, interpreting, and validating such bioenergetic tests (see also figure on the right).
Interlaboratory proficiency test
- Inter-laboratory ring tests are a basic requirement for quality management. Such proficiency tests require a reference sample which must be homogenous, stable, representative of the diagnostic target, amenable for distribution, and economical for large-scale production. For respiratory OXPHOS analysis a reference sample of structurally and functionally intact mitochondria is not yet available to perform corresponding ring tests (in contrast to enzymatic OXPHOS ring tests). Recently, the widely applied human cell line HEK 293T was found to be potentially suitable for cryopreservation. Upon thawing the cells can be used immediately for respirometry of both intact and permeabilized cells. Thus interlaboratory proficiency testing may now be feasible as a world-wide innovation in the field of mitochondrial respiratory physiology. Participating labs may implement the test for intra- and interlaboratory validation and longitudinal performance monitoring, using a reference OXPHOS protocol (Fig. 3). The organization of the proficiency test should follow the requirements of the ISO 17043 and 13528 standards. A corresponding MitoEAGLE proficiency training shall be developed and implemented. Feedback from the participants will lead to a final adjustment of the SOPs in the proficiency test, which will be made generally available.
Instrumental platforms
- Comparison of results between and within instrumental platforms: The reference protocols and proficiency test will not define the instrumental platform, but will allow a quantitative comparison of results obtained with specific equipment available in the participating laboratories.
Nomenclature
- Harmonization of nomenclature on mitochondrial respiratory states and control parameters: The logistics of development of a database requires application of strictly defined terms for all included variables. There is no general reference available upon which a consistent terminology on mitochondrial physiology and bioenergetics can be based. The MitoEAGLE consortium, therefore, has to accomplish the ambitious goal to unify and simplify the terminology in the field for the purpose of the QMS, which will lead to the development of recommendations for the use of a common terminology in mitochondrial physiological research. A publication will be prepared as an Open Access article which will be a milestone towards a unification of concepts and nomenclature.
Knowledge management system
- With an increase in data volume and complexity comes along the requirement for standardized report formats and digital accessibility of the data. Since such a database is still missing for mitochondrial physiology, the goal of WG1 is to establish an integrated and analytic environment linking mitochondrial respiration with external data (cell physiology, anthropometry, spiroergometry, lifestyle, genomics) in the context of physical fitness evaluation and clinical diagnostics. Databases will be established combining biological and clinical data comprising proprietary and publicly available datasets into one integrated solution. Data import and export functionalities will allow data upload and mapping to corresponding objects in the database, in this way generating valuable data and allowing for project-specific as well as cross-project data mining.
- The ability to formulate meta-studies in our approach is an important success factor: The platform can aggregate the project’s complete knowledge and provides the ability to make diagnostic profiling associated with diseases. In addition to functional data, other data types such as nutritional or environmental factors associated with disease prevention can be identified and aggregated into the MitoEAGLE Knowledge Management System.
- Querying the heterogeneous data types and result representation are the most important requirements of the knowledge platform besides to data integration and modelling. The application will feature different types of visualization techniques for all integrated data types. The database will be developed using the concept of multi-tier client-server architecture. Individual components are separated corresponding to their functionality. An upstream web server handles user requests, an application server performing business logic, and a back-end database system guaranties persistent data storage. All components are additionally separated by means of network security incorporating firewalls and are hosted in a secure environment. The platform administration ensures privacy regarding proprietary data by user rights policies.
- It will be the task of WG1 to continuously collect the input from all other WGs, compare, discuss and align SOPs as far as possible, and to finally develop recommendations for quality control, data reporting and data sharing beyond the published record. In addition, WG1 will develop concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation. Finally, it will provide a summary on strategic dissemination and an education programme for MitoEAGLE. The education programme will ensure that not only presently participating groups will adhere to the set standards but that also groups joining in at later points may smoothly fit into the community and help to further broaden and deepen our understanding on mitochondrial physiology in health and disease.
Management
Tasks
- Opening of the database to the research public for data search and moderated entry of new datasets.
- Development of interlaboratory proficiency ring test, of recommendation/certification reports and of a MitoEAGLE proficiency training module.
- Sample distribution, ring test experiments (MitoEAGLE-proficiency training on two different points of time, 6 months apart), data analysis by experimenters and MitoEAGLE-proficiency test manager.
- Reports and publication: Reports with specific results in comparison with an anonymous data summary, and joint publication. Individual reports may be complemented by recommendations, and are summarized in an informal certification report.
- Instrumental platform comparison with reference sample applying comparable measurements.
- Draft, dissemination, collection and final formulation of a manuscript on concepts and terminology of mitochondrial physiology.
- Collect and discuss procedures and experimental protocols for the evaluation of mitochondrial capacities and create a library of protocols: A database comprising reference SUIT protocols, standard experimental media, and detailed instructions for OXPHOS analysis is assembled.
- MitoEAGLE data management system (DMS) development: The DMS for further data preprocessing. The DMS will enable management and analysis of instrument-specific and project-specific data.
- MitoEAGLE database (DB) development: DB will facilitate high-level project specific analysis as well as meta-analyses across projects, visualization of project specific data as well as integrative analyses of datasets for various proprietary and/or publicly available datasets.
- Testing of developed software components regarding functionality and usability, modification and implementation. Deployment for routine testing in the labs of our strategic partners.
- Data input: Upload of datasets generated through WG 2-4 in internal lab developments, results of the proficiency ring test, and during application module developments. In addition, the database will be populated with available proprietary and public datasets.
- Develop recommendations for quality control, data reporting and data sharing beyond the published record. Develop concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation.
- Provide a summary on strategic dissemination and an education programme for MitoEAGLE.
Milestones
- Data sets entered in standardized format and database opened for the public
- Protocols for ring-tests and standard analysis and report scheme defined
- MitoEAGLE proficiency training workshop finished
- Analysis is completed and joint publication submitted
- Publication of paper on concepts and terminology
- Library of protocols online
- Publication of recommended protocols and procedures
- Guidelines for acquisition, evaluation and data documentation available
- MitoEAGLE data management system finished
- MitoEAGLE database ready for use
- Software modifications incorporated following extensive tests
- Publication of MitoEAGLE-KMP
- Recommendations on data acquisition, evaluation and management published
- Concepts on Open Access and institutionalized service for data management, data mining and health-care conforming standards of data interpretation published
Deliverables
- Guidelines for future research and recommendations for the evaluation of respiratory characteristics in human respirometric samples
- A database on mt fitness evaluated with human cells and tissues
- Training of researchers towards improved reproducibility of sample preparation and respirometric evaluation
- Qualitative and quantitative evaluation of results obtained in ongoing studies by comparison with a reference sample
- An expanded MitoEAGLE database
- A joint publication documenting the results of the MitoEAGLE-PT
- A multi-authored publication towards a unification of concepts and nomenclature in mitochondrial physiology
- A library of protocols for sample preparation and examination by respirometry
- A multi-authored publication with recommendations of comparable standard protocols and procedures.
- Guidelines as to the optimum use of a reference sample for determination of respiration and related parameters of interest.
- The MitoEAGLE data management system.
- A continuously updated MitoEAGLE database for the use as reference data and guideline towards application of optimized protocols, ultimately leading towards standardization.
- A joint publication on the use of the MitoEAGLE database and management system.
- An education programme for the use of MitoEAGLE database and management system.