Facucho-Oliveira 2009 Stem Cell Rev Rep
| Facucho-Oliveira JM, St John JC (2009) The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep 5:140-58. https://doi.org/10.1007/s12015-009-9058-0 |
Facucho-Oliveira JM, St John JC (2009) Stem Cell Rev Rep
Abstract: Pluripotent blastomeres of mammalian pre-implantation embryos and embryonic stem cells (ESCs) are characterized by limited oxidative capacity and great reliance on anaerobic respiration. Early pre-implantation embryos and undifferentiated ESCs possess small and immature mitochondria located around the nucleus, have low oxygen consumption and express high levels of glycolytic enzymes. However, as embryonic cells and ESCs lose pluripotency and commit to a specific cell fate, the expression of mtDNA transcription and replication factors is upregulated and the number of mitochondria and mtDNA copies/cell increases. Moreover, upon cellular differentiation, mitochondria acquire an elongated morphology with swollen cristae and dense matrices, migrate into wider cytoplasmic areas and increase the levels of oxygen consumption and ATP production as a result of the activation of the more efficient, aerobic metabolism. Since pluripotency seems to be associated with anaerobic metabolism and a poorly developed mitochondrial network and differentiation leads to activation of mitochondrial biogenesis according to the metabolic requirements of the specific cell type, it is hypothesized that reprogramming of somatic cells towards a pluripotent state, by somatic cell nuclear transfer (SCNT), transcription-induced pluripotency or creation of pluripotent cell hybrids, requires acquisition of mitochondrial properties characteristic of pluripotent blastomeres and ESCs.
• Bioblast editor: Gnaiger E
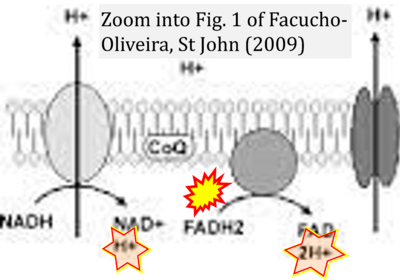
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.